Which 3 Enzymes Fix And Repair Damaged Dna
Deoxyribonucleic acid integrity is ever under attack from environmental agents like skin cancer-causing UV rays. How do Dna repair mechanisms detect and repair damaged Deoxyribonucleic acid, and what happens when they neglect?

Because DNA is the repository of genetic information in each living prison cell, its integrity and stability are essential to life. DNA, all the same, is not inert; rather, it is a chemical entity subject area to assault from the environs, and any resulting damage, if not repaired, will pb to mutation and possibly disease. Perhaps the best-known instance of the link between ecology-induced DNA damage and disease is that of pare cancer, which can be caused past excessive exposure to UV radiation in the course of sunlight (and, to a lesser degree, tanning beds). Some other example is the damage acquired past tobacco smoke, which tin can lead to mutations in lung cells and subsequent cancer of the lung. Across environmental agents, Dna is as well subject to oxidative impairment from byproducts of metabolism, such as free radicals. In fact, it has been estimated that an individual cell can suffer up to one million DNA changes per mean solar day (Lodish et al., 2005).
In addition to genetic insults acquired by the surroundings, the very procedure of DNA replication during prison cell division is prone to error. The rate at which Deoxyribonucleic acid polymerase adds incorrect nucleotides during DNA replication is a major gene in determining the spontaneous mutation charge per unit in an organism. While a "proofreading" enzyme normally recognizes and corrects many of these errors, some mutations survive this process. Estimates of the frequency at which human DNA undergoes lasting, uncorrected errors range from 1 10 x-4 to 1 x ten-vi mutations per gamete for a given gene. A rate of 1 x x-6 ways that a scientist would expect to find one mutation at a specific locus per one million gametes. Mutation rates in other organisms are often much lower (Table 1).
One fashion scientists are able to estimate mutation rates is by considering the rate of new dominant mutations found at different loci. For example, by examining the number of individuals in a given population who were diagnosed with neurofibromatosis (NF1, a illness caused by a spontaneous—or noninherited—dominant mutation), scientists determined that the spontaneous mutation rate of the gene responsible for this disease averaged 1 x 10-4 mutations per gamete (Crowe et al., 1956). Other researchers accept institute that the mutation rates of other genes, like that for Huntington's illness, are significantly lower than the rate for NF1. The fact that investigators have reported different mutation rates for unlike genes suggests that certain loci are more decumbent to damage or mistake than others.
Deoxyribonucleic acid Repair Mechanisms and Homo Disease

Deoxyribonucleic acid repair processes be in both prokaryotic and eukaryotic organisms, and many of the proteins involved have been highly conserved throughout development. In fact, cells take evolved a number of mechanisms to detect and repair the various types of damage that can occur to Dna, no matter whether this damage is caused past the environs or by errors in replication. Because Deoxyribonucleic acid is a molecule that plays an active and critical role in cell partitioning, control of Dna repair is closely tied to regulation of the cell cycle. (Recall that cells transit through a cycle involving the G1, S, G2, and M phases, with Dna replication occurring in the South phase and mitosis in the M phase.) During the cell wheel, checkpoint mechanisms ensure that a jail cell's DNA is intact before permitting DNA replication and cell segmentation to occur. Failures in these checkpoints tin can lead to an accumulation of damage, which in turn leads to mutations.
Defects in Dna repair underlie a number of human genetic diseases that bear upon a wide variety of body systems but share a constellation of common traits, near notably a predisposition to cancer (Table 2). These disorders include ataxia-telangiectasia (AT), a degenerative motor condition caused past failure to repair oxidative damage in the cerebellum, and xeroderma pigmentosum (XP), a status characterized by sensitivity to sunlight and linked to a defect in an important ultraviolet (UV) damage repair pathway. In improver, a number of genes that take been implicated in cancer, such equally the RAD group, have also been determined to encode proteins disquisitional for Dna damage repair.
UV Damage, Nucleotide Excision Repair, and Photoreactivation

As previously mentioned, one important DNA impairment response (DDR) is triggered past exposure to UV light. Of the iii categories of solar UV radiation, but UV-A and UV-B are able to penetrate World'south atmosphere. Thus, these ii types of UV radiation are of greatest concern to humans, particularly as continuing depletion of the ozone layer causes higher levels of this radiation to reach the planet's surface.
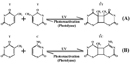
UV radiation causes ii classes of Dna lesions: cyclobutane pyrimidine dimers (CPDs, Figure one) and half-dozen-iv photoproducts (6-4 PPs, Figure 2). Both of these lesions misconstrue DNA's structure, introducing bends or kinks and thereby impeding transcription and replication. Relatively flexible areas of the DNA double helix are nearly susceptible to damage. In fact, one "hot spot" for UV-induced impairment is found within a commonly mutated oncogene, the p53 gene.
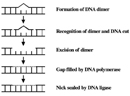
CPDs and six-4 PPs are both repaired through a process known every bit nucleotide excision repair (NER). In eukaryotes, this complex process relies on the products of approximately 30 genes. Defects in some of these genes have been shown to cause the human affliction XP, likewise every bit other weather condition that share a take chances of skin cancer that is elevated almost a thousandfold over normal. More specifically, eukaryotic NER is carried out past at least 18 protein complexes via 4 discrete steps (Figure 3): detection of damage; excision of the section of DNA that includes and surrounds the error; filling in of the resulting gap past Deoxyribonucleic acid polymerase; and sealing of the nick betwixt the newly synthesized and older DNA (Figure 4). In bacteria (which are prokaryotes), however, the procedure of NER is completed by only iii proteins, named UvrA, UvrB, and UvrC.
Bacteria and several other organisms likewise possess another mechanism to repair UV damage called photoreactivation. This method is oft referred to as "lite repair," because it is dependent on the presence of light free energy. (In comparison, NER and most other repair mechanisms are frequently referred to as "dark repair," as they do not crave light equally an free energy source.) During photoreactivation, an enzyme called photolyase binds pyrimidine dimer lesions; in addition, a second molecule known as chromophore converts lite free energy into the chemic free energy required to direct revert the afflicted area of DNA to its undamaged course. Photolyases are found in numerous organisms, including fungi, plants, invertebrates such as fruit flies, and vertebrates including frogs. They do not appear to exist in humans, still (Sinha & Hader, 2002).
Additional DNA Repair mechanisms
NER and photoreactivation are not the just methods of DNA repair. For case, base of operations excision repair (BER) is the predominant mechanism that handles the spontaneous Deoxyribonucleic acid damage caused by free radicals and other reactive species generated by metabolism. Bases can become oxidized, alkylated, or hydrolyzed through interactions with these agents. For example, methyl (CH3) chemic groups are frequently added to guanine to class 7-methylguanine; alternatively, purine groups may be lost. All such changes result in abnormal bases that must be removed and replaced. Thus, enzymes known equally DNA glycosylases remove damaged bases by literally cutting them out of the Dna strand through cleavage of the covalent bonds between the bases and the sugar-phosphate backbone. The resulting gap is then filled past a specialized repair polymerase and sealed by ligase. Many such enzymes are found in cells, and each is specific to certain types of base of operations alterations.
Yet some other form of Dna impairment is double-strand breaks, which are caused by ionizing radiation, including gamma rays and Ten-rays. These breaks are highly deleterious. In addition to interfering with transcription or replication, they can pb to chromosomal rearrangements, in which pieces of one chromosome become attached to some other chromosome. Genes are disrupted in this process, leading to hybrid proteins or inappropriate activation of genes. A number of cancers are associated with such rearrangements. Double-strand breaks are repaired through one of 2 mechanisms: nonhomologous cease joining (NHEJ) or homologous recombination repair (HRR). In NHEJ, an enzyme chosen DNA ligase Four uses overhanging pieces of DNA side by side to the intermission to join and fill in the ends. Additional errors tin can be introduced during this process, which is the instance if a cell has not completely replicated its DNA in grooming for sectionalisation. In contrast, during HRR, the homologous chromosome itself is used equally a template for repair.
Mutations in an organism's Dna are a part of life. Our genetic code is exposed to a diversity of insults that threaten its integrity. But, a rigorous system of checks and balances is in place through the DNA repair machinery. The errors that slip through the cracks may sometimes be associated with disease, but they are also a source of variation that is acted upon by longer-term processes, such as evolution and natural selection.
References and Recommended Reading
Branze, D., & Foiani, M. Regulation of Dna repair throughout the prison cell cycle. Nature Reviews Molecular Jail cell Biology ix, 297–308 (2008) doi:ten.1038/nrm2351.pdf (link to article)
Crowe, F. Westward., et al. A Clinical, Pathological, and Genetic Study of Multiple Neurofibromatosis (Springfield, Illinois, Charles C. Thomas, 1956)
Lodish, H., et al. Molecular Biology of the Jail cell, fifth ed. (New York, Freeman, 2004)
Sinha, R. P., & Häder, D. P. UV-induced Dna damage and repair: A review. Photochemical and Photobiological Sciences 1, 225–236 (2002)
Source: https://www.nature.com/scitable/topicpage/dna-damage-repair-mechanisms-for-maintaining-dna-344/
Posted by: eckerttoop1970.blogspot.com


0 Response to "Which 3 Enzymes Fix And Repair Damaged Dna"
Post a Comment